Exploring the relationship between geographic location and gut microbiome composition with classifiers
August 2023, Blekhman Lab
Abstract
The human gut microbiome, an intricate ecosystem of trillions of microorganisms, has been emphasized in its implications for human health and disease. While past research has uncovered its influences in ethnicity and socioeconomic status, the impact geography might hold has yet to be interrogated despite the noticeable bias in gathered data towards North America and Europe. Thus, we sought to identify whether one’s gut microbiome profile could be used to identify their geographical origins to assess whether there were significant composition differences across regions. To do this, we explored the performance of seven models, including deep neural networks, gradient boosting machines, and ensemble methods. As a preliminary investigation, our findings showed high variance and poor performance, underscoring potentially unknown complexities of the gut microbiome's association with geography. An inherent bias toward Europe and North America in the dataset highlights the need for better global representation. Future efforts should address class imbalance, experiment with more advanced architectures, and consider feature engineering methods for improved accuracy and determining the nature of the relationship.
Background
The human microbiome, comprising trillions of microorganisms like bacteria, fungi, parasites, and viruses, constitutes a dynamic ecosystem within the intestines. Microbiota, the constituents of the microbiome, play essential roles in stimulating the immune system, facilitating food breakdown, and synthesizing vital nutrients (Harvard School of Public Health, 2022).
Initiated during birth and influenced by factors such as diet and environment, an individual's microbiome undergoes continuous modifications, potentially affecting health outcomes. In a healthy state, the gut microbiota have myriad positive functions, including energy recovery from metabolism of nondigestible components of foods, protection of a host from pathogenic invasion, and modulation of the immune system. A dysbiotic state of the gut microbiota is becoming recognized as an environmental factor that interacts with a host’s metabolism and has a role in pathological conditions, both systemic—obesity, diabetes, and atopy—and gut-related IBS and IBD (Bull & Plummer, 2014). Studying the gut microbiome could provide insights into the diagnosis and functioning of disease and ways to optimize human health.
Recent work found that publicly available data on the human microbiome is heavily biased in favor of Europe and Northern America (Abdill et al. 2022). Since much of what we know regarding the links between microbiome composition and disease is based on studying populations in this region, we sought to examine whether the microbiomes of people elsewhere in the world differed consistently enough that a classifier could determine whether a single sample originated from Europe or North America.
Gut microbiome classification has had several breakthroughs over recent years. Scientists have have identified ethnicity (Dwiyanto et al., 2021, Brooks et al., 2018) and even socioeconomic status (Ahn et al., 2023) as factors that influence one’s microbiota. However, little research has been done to uncover the impact geographical location may hold as well. Here, we explored this by seeing if one’s continent of origin could be accurately identified with just their gut profile. Several machine learning models were trialed to interrogate the potential existence of this relationship.
Results
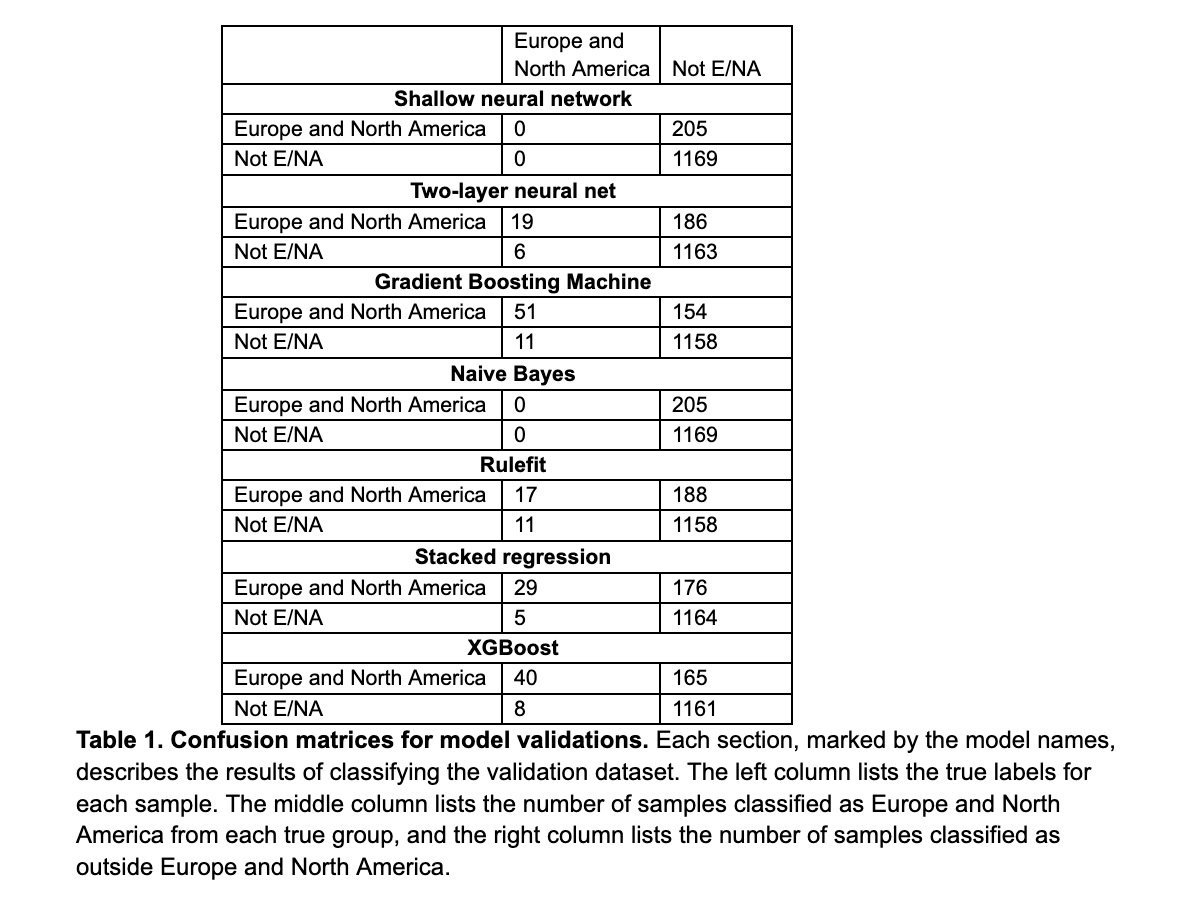
With an eye to eventually developing a classifier capable of inferring the geographic origin of a human gut microbiome sample, we performed a pilot study in which we evaluated the efficacy of seven models trained on a set of 7,000 samples of publicly available 16S rRNA amplicon sequencing data. Given the overrepresentation of Europe and North America in the world's largest sequencing data repositories (Abdill et al. 2022), we chose a binary classification task in which the outcome was whether the sample originated from outside of Europe and North America. In all cases, we observed high variance and poor performance on validation data. For each of the tests below, we used the same 80/20 split of the 7,000 samples, and the validation dataset was made of up 1,374 samples—205 from Europe and North America, and 1,169 from other regions of the world.
Our first model was a shallow neural network with a single hidden node that achieved about 85 percent accuracy—unfortunately, this is an artifact of the class imbalance in the data, and the classifier assigned all validation samples to the "Not Europe and Northern America" category, a result we also observed when training a naive Bayes classifier. Expanding this neural network to two layers, of 200 and 100 neurons respectively, yielded marginal improvement to overall accuracy, but still resulted in a 90.7 percent error rate for classifying samples from Europe and North America.
Next, we trained a gradient boosting machine, which correctly classified more European samples (51) than any other, but even this resulted in 82.3 percent precision and only 24.9 percent recall. We were optimistic about applying the H2O library's RuleFit algorithm to this dataset, though this too struggled to classify the samples and returned a 91.7 percent error rate for European samples.
A stacked regression approach yielded 85.3 percent precision in detecting European samples, but still resulted in only 14.2 percent recall. An XGBoost-based algorithm was similarly unsuccessful, with 83.3 percent precision and 19.5 percent recall.
These results suggest that it is challenging to determine the geographic origin of a human gut microbiome sample. The observed high variance and poor performance across various models highlight the complexity of the relationship between gut microbiome composition and geographic location.
One key observation is the class imbalance in the data, with overrepresentation from Europe and North America. This imbalance likely contributed to skewed results, as seen in the shallow neural network and naive Bayes classifier, which exhibited high accuracy but failed to correctly classify European samples. This inherent geographical bias in our dataset could have contributed to the poor performance of our models, emphasizing the need for more equitable representation in microbiome sampling.
Moreover, the marginal improvement with a two-layer neural network and the mixed performance of gradient boosting and RuleFit algorithms indicate that the relationship between gut microbiome and geographic location may not be linear. The stacked regression approach and XGBoost-based algorithm demonstrated some precision but lacked recall, indicating a struggle to identify European samples comprehensively. This suggests that the models may need to be more sensitive to the nuances of microbiome data, especially when dealing with diverse global populations.
Next steps could involve refining the dataset by ensuring better representation from underrepresented regions, thereby mitigating class imbalance. Additionally, experimenting with more advanced models, such as more complex deep learning architectures with varied hyperparameters, ensemble methods, or attention mechanisms, might capture the intricate patterns in the microbiome data that contribute to geographic distinctions. Feature engineering techniques could also be useful for classification or worthwhile for other analyses if specific taxa or combinations of taxa were found to correlate with certain continents of origin. In such future work, unsupervised learning methods like clustering could provide insight.
Methods
The dataset (Abdill et al. 2022) was initially sorted by the continent of origin for each sample, so the first step was to shuffle the dataset with Pandas. This was done to ensure that, upon splitting the data into training and validation subsets, each would receive an even distribution of continents. 80% of the dataset was assigned for training and 20% was assigned for validation.
Two other preprocessing methods were then utilized before we began training our machine learning models. Firstly, taxa below 80% prevalence were removed, to reduce the size of the data by excluding comparatively rare taxa that were unlikely to be useful to the models. The robust center logratio (CLR) transformation was then applied to the dataset (Martino et al. 2019).
The machine learning models were implemented using Python's H2O library for training and validation. The deep neural network (DNN) employed a single hidden layer with a Tanh activation function. The DNN, implemented with the H2ODeepLearningEstimator function, was trained for 1000 epochs with a multinomial distribution, using parameters such as hidden=[1], activation="Tanh", and other settings for reproducibility and balance. The model's performance metrics, including accuracy for each class and area under the curve, were assessed on the validation dataset.
The gradient boosting machine (GBM) was implemented with the H2OGradientBoostingEstimator function, using a 5-fold cross-validation approach. Hyperparameters such as nfolds=5 and seed=1111 were specified. The Naive Bayes classifier, implemented with H2ONaiveBayesEstimator, utilized Laplace smoothing and 5-fold cross-validation with laplace=0 and seed=1234.
The RuleFit algorithm, implemented with H2ORuleFitEstimator, was trained with a maximum rule length of 10 and a maximum of 100 rules using parameters like max_rule_length=10 and max_num_rules=100. Performance metrics and predictions were recorded.
Additionally, a stacked ensemble algorithm was trained using a grid search over hyperparameters for the GBM. The ensemble, implemented with H2OStackedEnsembleEstimator, combined multiple GBM models with varying learning rates, maximum depths, sample rates, and column sample rates. The grid search was performed using parameters such as hyper_params and search_criteria. The ensemble's performance was compared to individual base models on the validation dataset.
Finally, an extreme gradient boost (XGBoost) algorithm with the dart booster algorithm was implemented using H2OXGBoostEstimator. Hyperparameters like booster='dart' and normalize_type="tree" were specified. The XGBoost model's performance metrics, predictions, and feature interactions were recorded.
Guides for how to reproduce each model in H2O can be found within their Python documentation ("H2O," n.d.).
References
Abdill, R. J., Adamowicz, E. M., & Blekhman, R. (2022). Public human microbiome data are dominated by highly developed countries. PLOS Biology, 20(2). https://doi.org/10.1371/journal.pbio.3001536
Ahn, J., Kwak, S., Usyk, M., Beggs, D., Choi, H., Ahdoot, D., Wu, F., Maceda, L., Li, H., Im, E. O., Han, H. R., Lee, E., Wu, A., & Hayes, R. (2023). Sociobiome - Individual and neighborhood socioeconomic status influence the gut microbiome in a multi-ethnic population in the US. Research square, rs.3.rs-2733916. https://doi.org/10.21203/rs.3.rs-2733916/v1
Brooks, A. W., Priya, S., Blekhman, R., & Bordenstein, S. R. (2018). Gut microbiota diversity across ethnicities in the United States. PLoS biology, 16(12), e2006842. https://doi.org/10.1371/journal.pbio.2006842
Bull, M. J., & Plummer, N. T. (2014). Part 1: The Human Gut Microbiome in Health and Disease. Integrative medicine (Encinitas, Calif.), 13(6), 17–22.
Dwiyanto, J., Hussain, M.H., Reidpath, D. et al. Ethnicity influences the gut microbiota of individuals sharing a geographical location: a cross-sectional study from a middle-income country. Sci Rep 11, 2618 (2021). https://doi.org/10.1038/s41598-021-82311-3
H2O. (n.d.). H2O documentation. H2O Documentation - H2O documentation. https://docs.h2o.ai/h2o/latest-stable/h2o-py/docs/index.html
Kucera, M., & Malmgren, B. (1998). Logratio transformation of compositional data—a resolution of the constant sum constraint. Marine Micropaleontology, 34(2), 117-120.
Martino, C., Morton, J. T., Marotz, C. A., Thompson, L. R., Tripathi, A., Knight, R., & Zengler, K. (2019). A novel sparse compositional technique reveals microbial perturbations. mSystems, 4(1). https://doi.org/10.1128/msystems.00016-19